Single Cell Sequencing
The Emory NPRC Genomics Core is the proud owner of a state-of-the-art 10x Genomics Chromium X instrument. This device allows us to meet all your single cell sequencing needs with high throughput and accuracy. The 10x Chromium platform uses GEM-X technology to individually capture and barcode single cells or nuclei for downstream library prep and analysis. Each individual cell or nucleus is partitioned within a droplet containing a Gel Bead carrying the necessary adaptors and barcodes to label each cell in isolation for targeted analysis.

We offer full support (capture, library prep and sequencing) of the reverse transcription (RT)-based 3' or 5' assays for whole transcriptome profiling alongside multiomic options. The Core can perform captures targeting up to 20,000 cells or nuclei.
We provide several analysis options to fit the parameters of your experimental needs, and can perform many of these assays simultaneously (GEX, VDJ, CSP):
- Gene Expression (GEX) Library: Reveals clonality, cellular diversity, and heterogeneity of the captured cells.
- VDJ Library: Assembles full-length V(D)J gene sequences and pairs heavy and light chain Ig sequences (BCR) or TRA and TRB sequences (TCR) for individual cells at full isotype resolution. Only compatible with the 5' assay and human, mouse, or rhesus samples.
- Cell Surface Protein (CSP) Library (feature barcoding): Allows for the analysis of other cellular data such as cell surface protein expression and antigen specificity to further interrogate the complex interactions of immune cells. Compatible with both the 3' and 5' assays. For CSP, anti-human and anti-mouse barcoded antibodies are commercially available, where many of the antibodies in the human portfolio are cross-reactive with rhesus.
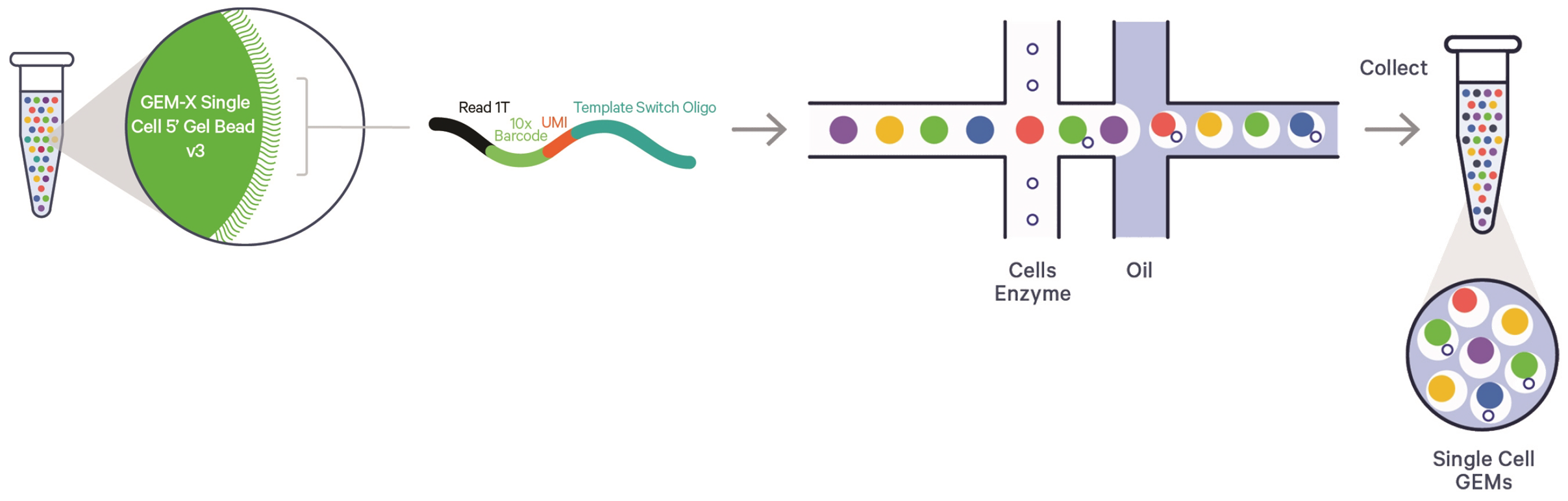
Image credit: 10x Genomics
The Genomics Core offers solutions for mapping the epigenetic landscape at single cell resolution. We support both the standalone Assay for Transposase Accessible Chromatin (ATAC-seq) workflow or the Multiomic (ATAC + 3' GEX analysis) workflow and can perform isolation of nuclei from cell suspensions or snap frozen tissue. The 10x Genomics Epi Chromatin assays currently have the capability to capture and analyze up to 10,000 nuclei.
- Epi ATAC, standalone ATAC Library only: Resolve cell types and states by exploring open chromatin regions and using genome-wide, epigenetic profiles.
- Epi Multiome ATAC + 3' Gene Expression (GEX) Library: Simultaneously profile gene expression and open chromatin from the same nucleus.
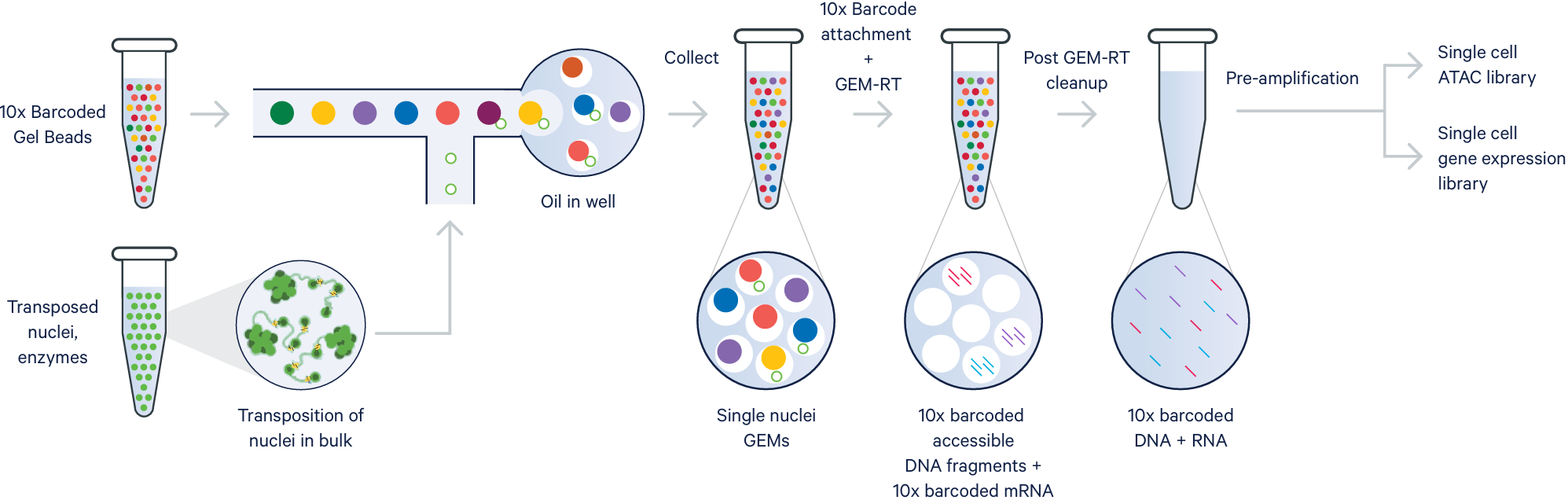
Image credit: 10x Genomics
The Flex Gene Expression assay provides an alternative solution for projects where there are logistical challenges in collecting and preparing freshly isolated samples for single cell capture. Fixation of samples with paraformaldehyde (PFA) at the point of collection preserves fragile cells and locks in the biological state, allowing storage or shipment of the samples and capture at a later date. This assay reduces experimental variability and increases the efficiency of captures by allowing strategic batching and multiplexing of samples. This workflow removes the time-sensitive nature of captures and allows researchers and the core to have increased flexibility.
The Flex Gene Expression assay is probe-based, offering whole transcriptome protein-coding gene coverage. This is compatible with human or mouse samples, fixed cells or nuclei, and there are different multiplexing options and the ability to include the analysis of cell surface protein. Multiplexing in the Flex assay is in-built, where the distinct probe set added to each sample has a unique probe set barcode, so no upstream labelling or tagging of cells is required.
- Flex Single-plex: one sample and up to 20,000 cells per channel/GEM reaction
- Flex 4-plex: up to four samples and 80,000 cells per channel/GEM reaction
- Flex 16-plex: up to sixteen samples and 320,000 cells per channel/GEM reaction
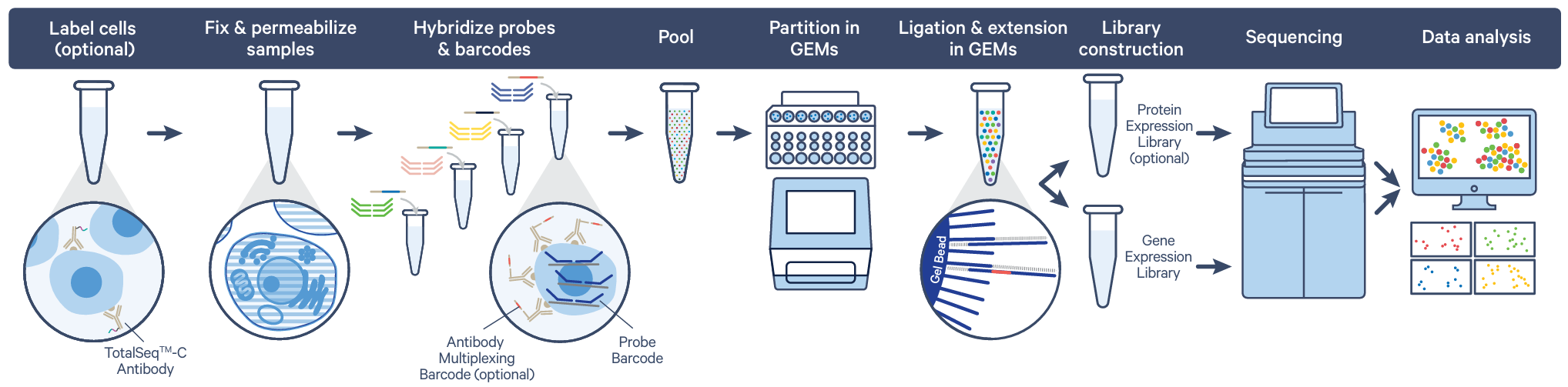
Image credit: 10x Genomics
There are two sample multiplexing options available for the Universal Gene Expression assays.
Hashtag oligos
Hashtags are a mixture of two monoclonal antibodies conjugated to the same feature barcode oligonucleotide. For mouse, the two antibodies are specific against CD45 and MHC class I, respectively, and for human, the two antibodies recognize CD298 and β2 microglobulin, respectively. These respective hashtag reagents can be used to label most cells in mice or humans.
Individual samples are stained with hashtag antibodies that have a distinct barcode sequence before they are pooled for capture. For hashtag antibodies bound to the cell surface protein, the feature barcode is directly captured by the Gel Bead oligo, amplified, and used for cell surface protein library construction. Demultiplexing is done bioinformatically, where cells are assigned to their sample through detection of the sample-specific hashtag barcode sequence.
This approach is limited to samples derived from mice, humans, and rhesus. We do not currently support hashing of nuclei.

Image credit: 10x Genomics
On-chip multiplexing
On-chip multiplexing (OCM) is facilitated by the design of the microfluidic chip in the Universal Gene Expression (3' or 5') 4-plex assay. This is accomplished by co-partitioning up to four samples from each multiplexing set with corresponding gel beads, each containing a unique list of 10x barcodes. A pool of GEMs is generated in a single recovery well for each multiplexing set. During data analysis, the 10x barcodes are used to computationally demultiplex the samples.
Upstream labelling or tagging of cells is not required. This approach is species agnostic and compatible with nuclei.
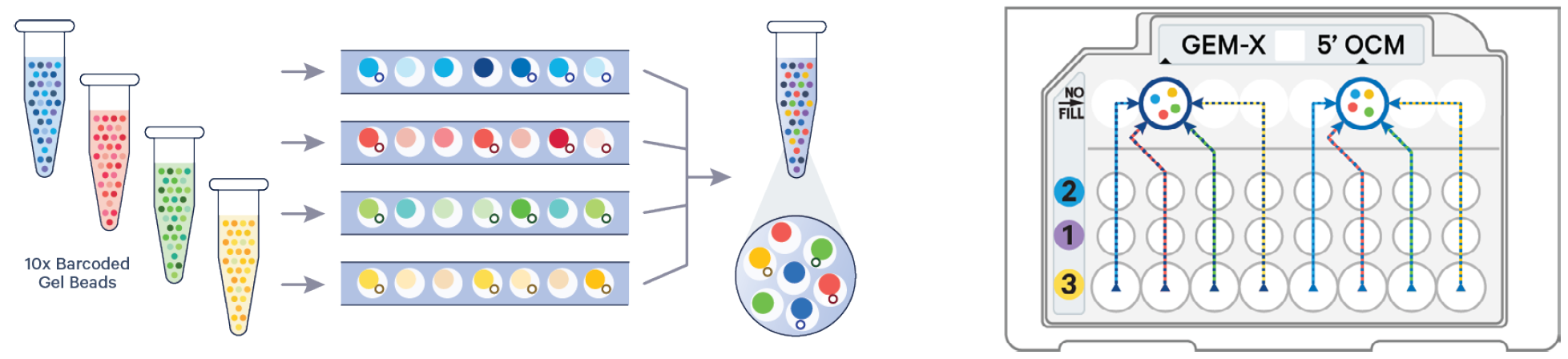
Image credit: 10x Genomics