Spatial Transcriptomics Projects
Spatial transcriptomics projects can be logistically challenging. Please contact the Genomics Core as soon as you start planning your experiments. The initial tissue sectioning/placement, staining and imaging steps may need to be completed by the appropriate histology/imaging core, but we will help you to loop them in after you contact us. The workflow is time sensitive and tight coordination of the different components is required.
Please review the considerations for the Visium HD assay below which are dependent on the tissue storage type and desired staining method.
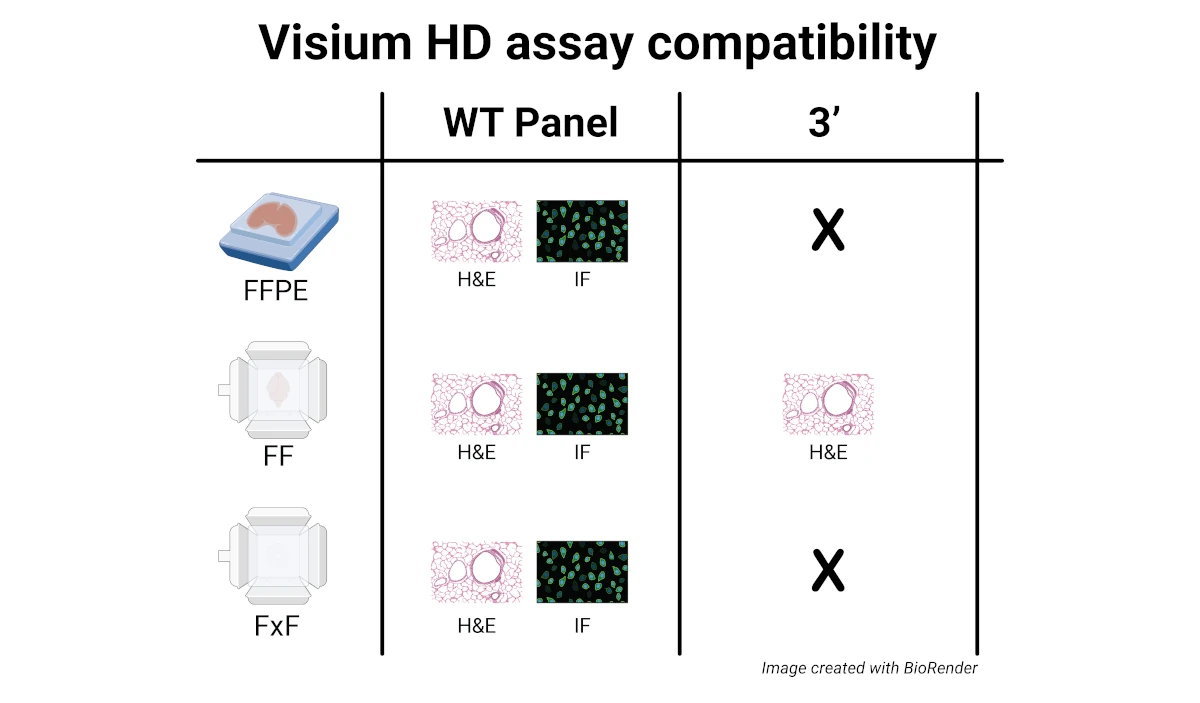
Formalin-Fixed, Paraffin Embedded (FFPE) tissue blocks
- Slides with freshly placed tissue sections prepared specifically for Visium HD are preferred
- Archived slides (H&E stained only, prepared with a hardset coverslip, imaged and stored) are compatible with the Visium HD assay but not recommended, as the tissues on these slides may have experienced RNA degradation over time.
- RNA quality assessment of the tissue in the FFPE block must be performed first. The recommended DV200 is >30%, where lower scores will likely impact assay performance. DV200 is the percentage of RNA fragments in the sample that are >200 nucleotides in length.
- Hematoxylin & Eosin (H&E) staining - the full workflow is supported by the EPC Histology and Molecular Pathology Laboratories or CTPSR and the Genomics Core (workflow is shown in flowchart)
- Immunofluorescence (IF) staining - the EPC Histology and Molecular Pathology Laboratories and CTPSR can help with the sectioning and tissue placement on glass slides, but the IF staining and imaging portions of the workflow must be completed yourself. The IF staining workflow may require additional 10x Genomics accessories or reagents that must be obtained from or purchased by the Genomics Core. The steps downstream of coverslip removal are fully supported by the Genomics Core.
- Detailed information for the Visium HD FFPE workflows can be found in this guide.
Fresh-Frozen (FF) or Fixed Frozen (FxF) OCT-embedded tissue blocks
- RNA quality assessment of the tissue in the frozen blocks is required (DV200 for FxF or RIN for FF). For FxF, the DV200 should be > 50%. For FF, tissue sections with RIN ≥ 4 are optimal for the Visium HD WT assay, and tissue sections with RIN ≥ 7 are optimal for the Visium HD 3’ assay.
- Hematoxylin & Eosin (H&E) staining - all the upstream histology (sectioning, placement,
staining for H&E, imaging) must be performed yourself or with an alternative histology
core. The downstream library prep component is fully supported by the Genomics Core. - Immunofluorescence (IF) staining – all the upstream histology (sectioning, placement,
staining for IF, imaging) must be performed yourself or with an alternative histology core. The downstream library prep component is fully supported by the Genomics Core. - The FF or FxF workflows (H&E or IF) may require additional 10x Genomics accessories or reagents that must be obtained from or purchased by the Genomics Core.
- Detailed information for the Visium HD WT Panel FF, FxF or Visium HD 3’ FF workflows can be found in the linked guides.